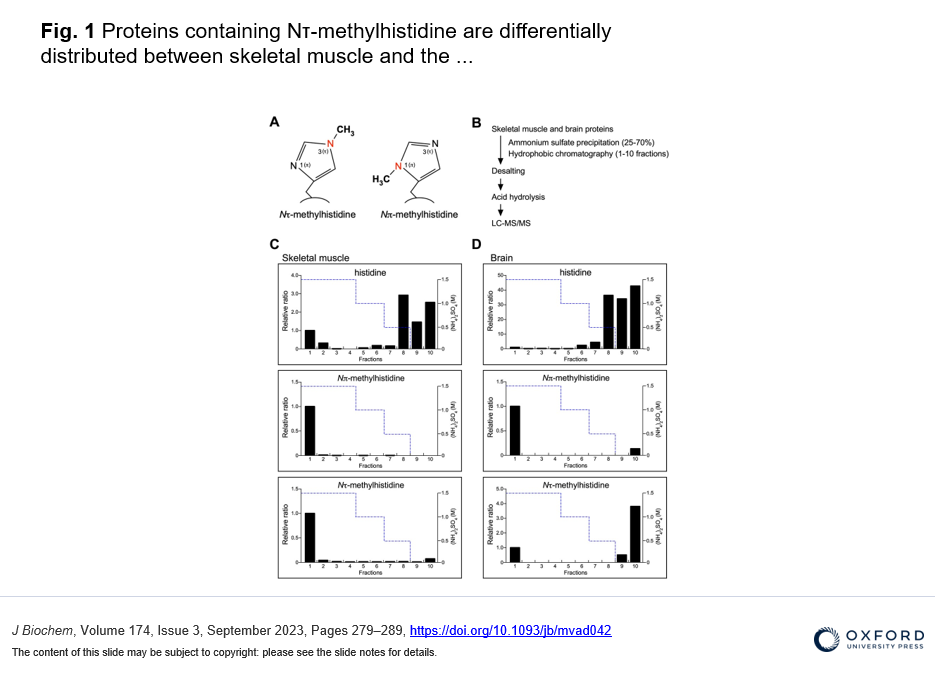
・The differential distribution pattern of Nτ-methylated proteins was found between the brain and skeletal muscle, and identified γ-enolase where the His-190 at the Nτ position is methylated in mouse brain.
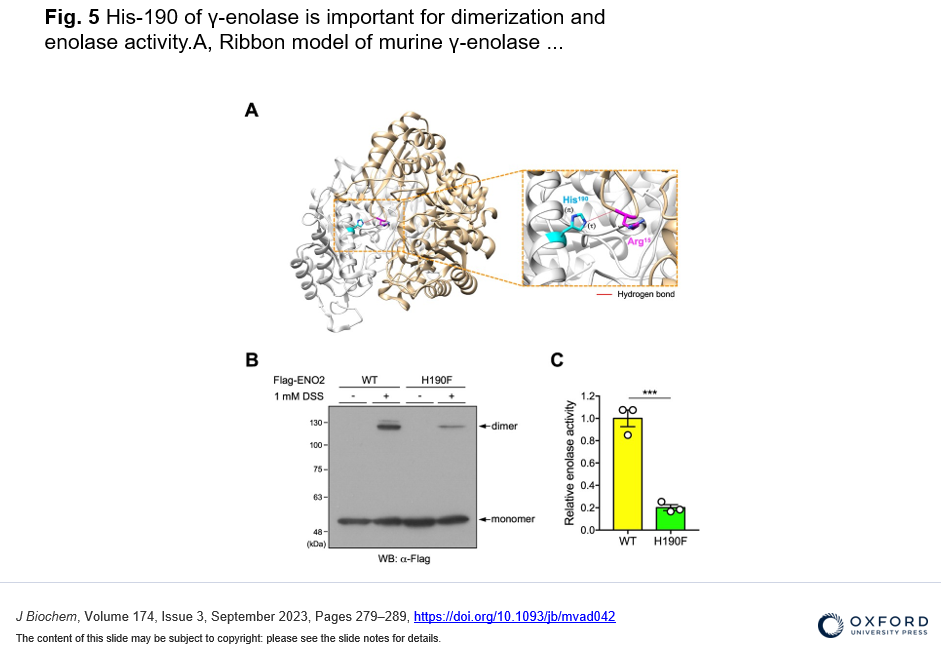
・In silico structural prediction and biochemical analysis showed that the His-190 in γ-enolase is involved in the intermolecular homodimeric formation and enzymatic activity.
・γ-enolase was methylated in brain tissue extracts, indicating the presence of a histidine methyltransferase other than the three known ones in mouse brain tissue.
Abstract
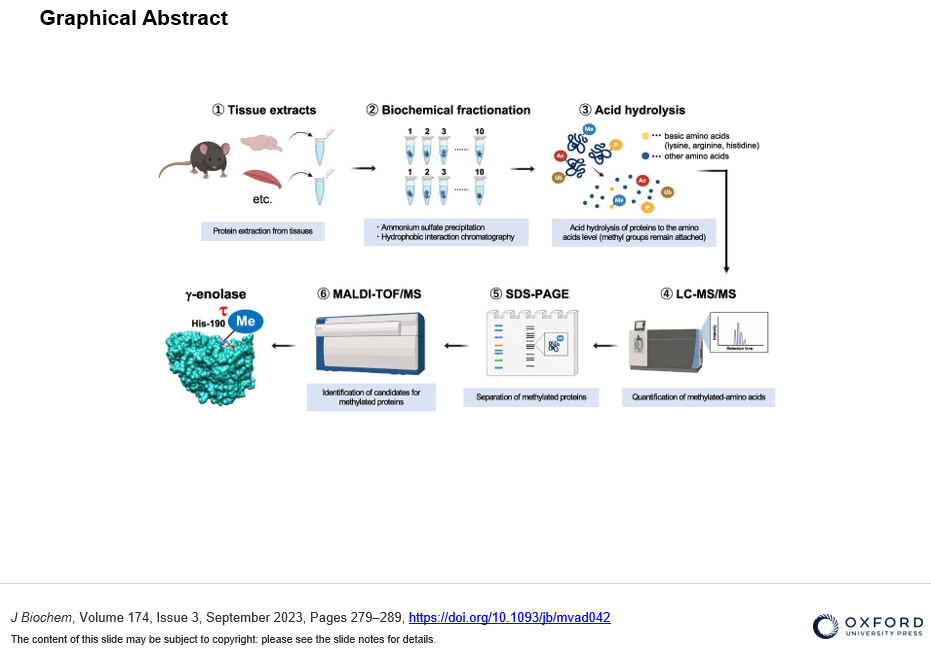
To identify new substrates for histidine methylation, we at first established a method to screen histidine-methylated proteins with the use of protein fractionation combined with subsequent quantitative analysis of acid hydrolysate followed by LC-MS/MS. Interestingly, using our techniques, we revealed that the distribution pattern of Nτ-methylated proteins in the brain was completely different from that of skeletal muscle. In addition, we identified γ-enolase, also known as ENO2 or neuron-specific enolase as a novel target protein of Nτ-methylation on His-190 in mouse brain. To investigate the role of His-190 in γ-enolase, we attempted to examine the position of this residue in the whole 3D structure using in silico structural analysis. Importantly, we found that this mutation significantly reduced the formation of the homodimeric γ-enolase and the enolase activity. These results showed that the histidine residue at 190 of γ-enolase is important for its homodimeric formation of γ-enolase and its enzymatic activity. Moreover, we performed in vitro methylation assay with the known methyltransferases SETD3, METTL9 and METTL18, but γ-enolase was not methylated by these enzymes. On the other hand, surprisingly, γ-enolase was methylated in brain tissue extracts, indicating the presence of a histidine methyltransferase other than the three known ones in mouse brain tissue.
Benefit
We provide a new methodology to find histidine-methylated proteins in vivo and identify y-enolase, a subunit of the glycolytic enzyme enolase, as a new substrate for methylation modification that occurs at histidine residues in proteins. We also identify the histidine residue where this methylation occurs and reveal that this is important for its homodimeric formation of γ-enolase and its enzymatic activity. We also indicate presence of a histidine methyltransferase other than the three known ones in mouse brain tissue.
Market Application
A method to identify new substrates by screenning histidine-methylated proteins with the use of protein fractionation combined with subsequent quantitative analysis of acid hydrolysate followed by LC-MS/MS. This methodology is advantageous in that the target protein can be explored, whether it contains Nπ- or Nτ-methylhistidine.
Publications
https://academic.oup.com/jb/article/174/3/279/7190349